To fill NaN values, pandas library itself provides a function fillna() to replace NaN values.
However there is an advanced Classes in scikit-learn which allows us to impute missing values.
It not only fills NaN values but also allows us to specify the placeholder which indicates the format of the missing value. It can be specified using the parameter “missing_values”.
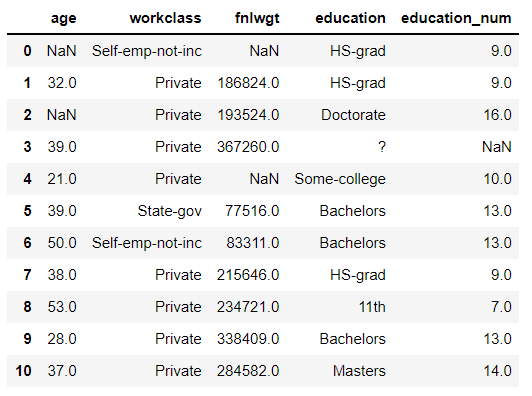
Data:
How to Impute NaN using SimpleImputer?
SimpleImputer is a transformer to impute missing values.
This works on entire data and it cannot be applied on a particular column.
For each missing value type, we have to define a separate imputer and fit-transform one by one. For example, all NaN can be imputed in one go and then to impute missing values mentioned as ‘?’ will have to be transformed in next call.
Allowed values: int, float, str, np.nan or None, default=np.nan
Disadvantages:
Different imputation cannot be applied for different columns.
Steps to use:
1. Import the Class.
from sklearn.impute import SimpleImputer
2. Define the Imputation by calling the SimpleImputer constructor.
We are creating an imputer ‘impute_mode_all‘ to replace all the NaN values with most frequent values.
But in our data, the column ‘education’ has a ‘?’ in place of the missing value. So we need to create another imputer for this, ‘impute_ques_all‘.
impute_mode_all = SimpleImputer(missing_values=np.nan, strategy='most_frequent')
impute_ques_all = SimpleImputer(missing_values='?', strategy='most_frequent')
3. Call fit-transform function to set data for imputation and transform.
After imputing the NaN values, we need to fit-transform the cleaned data with the imputer created for ‘?‘.
data_clean = impute_mode_all.fit_transform(nan_data)
data_clean = impute_ques_all.fit_transform(data_clean)
data_clean
array([[39.0, 'Self-emp-not-inc', 77516.0, 'HS-grad', 9.0], [32.0, 'Private', 186824.0, 'HS-grad', 9.0], [39.0, 'Private', 193524.0, 'Doctorate', 16.0], [39.0, 'Private', 367260.0, 'Bachelors', 9.0], [21.0, 'Private', 77516.0, 'Some-college', 10.0], [39.0, 'State-gov', 77516.0, 'Bachelors', 13.0], [50.0, 'Self-emp-not-inc', 83311.0, 'Bachelors', 13.0], [38.0, 'Private', 215646.0, 'HS-grad', 9.0], [53.0, 'Private', 234721.0, '11th', 7.0], [28.0, 'Private', 338409.0, 'Bachelors', 13.0], [37.0, 'Private', 284582.0, 'Masters', 14.0]], dtype=object)
I have changed the text color to red for the imputed values in the above result for easy understanding.
SimpleImputer returns the result in array format instead of DataFrame.
How to Impute NaN using ColumnTransformer?
Advantage:
ColumnTransformer allows transformation in different columns with different imputations and applies at the same time.
Steps to use:
1. Import the Class.
from sklearn.compose import ColumnTransformer
2. Define the Imputation by calling the SimpleImputer constructor.
Here we have created a mean imputer for column index 0 – age column, mode imputer for columns 3rd & 4th. Even in that we have mentioned the missing values as ‘?’ for education column.
Column transformer returns only the columns which were transformed by default. If we need to retain other non-imputed columns too in the result, then we need to mention the parameter remainder as ‘passthrough’. The column ‘workclass’ was not transformed. As we have set remainder parameter as ‘passthrough’, it is existing in result.
impute_mean = SimpleImputer(missing_values=np.nan, strategy='mean')
impute_mode = SimpleImputer(missing_values='?', strategy='most_frequent')
impute_mode_num = SimpleImputer(missing_values=np.nan, strategy='most_frequent')
column_trans = ColumnTransformer([('impute_age', impute_mode_num, [0]),
('impute_fnlwgt', impute_mean, [2]),
('impute_education', impute_mode, [3]),
('impute_edu_num', impute_mode_num, [4])],
remainder='passthrough')
3. Call fit-transform function to set data for imputation and transform.
column_names = data.columns
data = column_trans.fit_transform(data)
Similar to SimpleImputer, as SimpleImputer is working behind, the return value will be an array instead of dataframe. So convert it to dataframe.
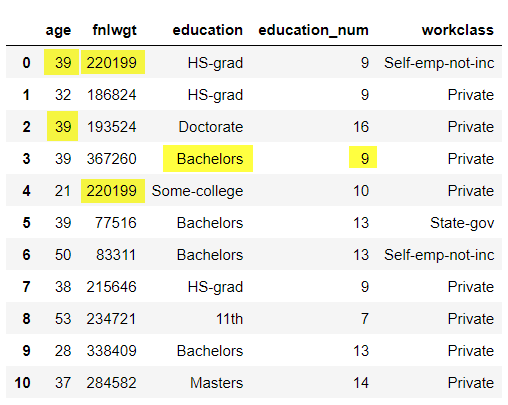
data_imputed = pd.DataFrame(data, columns = column_names).reset_index(drop='index')
data_imputed
Conclusion:
In this article we have learned imputation using the sklearn module SimpleImputer and ColumnImputer.
These 2 modules can be used for more pre-processing techniques such as categorical value encoding, data transformation and Standardization etc.
We will see all these usages in a different post.